Rabbit Anti-phospho-EP300 (Ser1834)antibody
EP300_HUMAN; Histone acetyltransferase p300; EC:2.3.1.48; p300 HAT; E1A-associated protein p300; Histone butyryltransferase p300; Histone crotonyltransferase p300; Protein 2-hydroxyisobutyryltransferase p300; Protein lactyltransferas p300; Protein propion
View History [Clear]
Details
Product Name phospho-EP300 (Ser1834) Chinese Name 磷酸化组蛋白乙酰转移酶p300抗体 Alias EP300_HUMAN; Histone acetyltransferase p300; EC:2.3.1.48; p300 HAT; E1A-associated protein p300; Histone butyryltransferase p300; Histone crotonyltransferase p300; Protein 2-hydroxyisobutyryltransferase p300; Protein lactyltransferas p300; Protein propionyltransferase p300; P300; KAT3B; MKHK2; RSTS2; literatures Product Type Phosphorylated anti Research Area Tumour immunology transcriptional regulatory factor Immunogen Species Rabbit Clonality Polyclonal React Species Human, Mouse, (predicted: Rat, Chicken, Dog, Pig, Cow, Horse, Guinea Pig, ) Applications WB=1:500-2000 ELISA=1:5000-10000 IHC-P=1:100-500 IHC-F=1:100-500 IF=1:100-500 (Paraffin sections need antigen repair)
not yet tested in other applications.
optimal dilutions/concentrations should be determined by the end user.Theoretical molecular weight 266kDa Cellular localization The nucleus cytoplasmic Form Liquid Concentration 1mg/ml immunogen KLH conjugated Synthesised phosphopeptide derived from human EP300 around the phosphorylation site of Ser1834: MA(p-S)MQ Lsotype IgG Purification affinity purified by Protein A Buffer Solution 0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol. Storage Shipped at 4℃. Store at -20 °C for one year. Avoid repeated freeze/thaw cycles. Attention This product as supplied is intended for research use only, not for use in human, therapeutic or diagnostic applications. PubMed PubMed Product Detail This gene encodes the adenovirus E1A-associated cellular p300 transcriptional co-activator protein. It functions as histone acetyltransferase that regulates transcription via chromatin remodeling and is important in the processes of cell proliferation and differentiation. It mediates cAMP-gene regulation by binding specifically to phosphorylated CREB protein. This gene has also been identified as a co-activator of HIF1A (hypoxia-inducible factor 1 alpha), and thus plays a role in the stimulation of hypoxia-induced genes such as VEGF. Defects in this gene are a cause of Rubinstein-Taybi syndrome and may also play a role in epithelial cancer. [provided by RefSeq, Jul 2008]
Function:
Functions as histone acetyltransferase and regulates transcription via chromatin remodeling. Acetylates all four core histones in nucleosomes. Histone acetylation gives an epigenetic tag for transcriptional activation. Mediates cAMP-gene regulation by binding specifically to phosphorylated CREB protein. Also functions as acetyltransferase for nonhistone targets. Acetylates 'Lys-131' of ALX1 and acts as its coactivator in the presence of CREBBP. Acetylates SIRT2 and is proposed to indirectly increase the transcriptional activity of TP53 through acetylation and subsequent attenuation of SIRT2 deacetylase function. Acetylates HDAC1 leading to its inactivation and modulation of transcription. Acts as a TFAP2A-mediated transcriptional coactivator in presence of CITED2. Plays a role as a coactivator of NEUROD1-dependent transcription of the secretin and p21 genes and controls terminal differentiation of cells in the intestinal epithelium. Promotes cardiac myocyte enlargement. Can also mediate transcriptional repression. Binds to and may be involved in the transforming capacity of the adenovirus E1A protein. In case of HIV-1 infection, it is recruited by the viral protein Tat. Regulates Tat's transactivating activity and may help inducing chromatin remodeling of proviral genes. Acetylates FOXO1 and enhances its transcriptional activity.
Subunit:
Interacts with phosphorylated CREB1. Interacts with HIF1A; the interaction is stimulated in response to hypoxia and inhibited by CITED2. Interacts (via N-terminus) with TFAP2A (via N-terminus); the interaction requires CITED2. Interacts (via CH1 domain) with CITED2 (via C-terminus). Interacts with CITED1 (unphosphorylated form preferentially and via C-terminus). Interacts with ESR1; the interaction is estrogen-dependent and enhanced by CITED1. Interacts with DTX1, EID1, ELF3, FEN1, LEF1, NCOA1, NCOA6, NR3C1, PCAF, PELP1, PRDM6, SP1, SP3, SPIB, SRY, TCF7L2, TP53, DDX5, DDX17, SATB1, SRCAP, TTC5, JMY and TRERF1. The TAZ-type 1 domain interacts with HIF1A. Probably part of a complex with HIF1A and CREBBP. Part of a complex containing CARM1 and NCOA2/GRIP1. Interacts with ING4 and this interaction may be indirect. Interacts with ING5. Interacts with the C-terminal region of CITED4. Interacts with HTLV-1 Tax and p30II. Interacts with and acetylates HIV-1 Tat. Non-sumoylated EP300 preferentially interacts with SENP3. Interacts with SS18L1/CREST. Interacts with ALX1 (via homeobox domain). Interacts with NEUROD1; the interaction is inhibited by NR0B2. Interacts with TCF3. Interacts (via CREB-binding domain) with MYOCD (via C-terminus). Binds to HIPK2. Interacts with ROCK2 and PPARG. Forms a complex made of CDK9, CCNT1/cyclin-T1, EP300 and GATA4 that stimulates hypertrophy in cardiomyocytes. Interacts with IRF1 and this interaction enhances acetylation of p53/TP53 and stimulation of its activity. Interacts with FOXO1; the interaction acetylates FOXO1 and enhances its transcriptional activity. Interacts with DDIT3/CHOP.
Subcellular Location:
Cytoplasm. Nucleus. Note=In the presence of ALX1 relocalizes from the cytoplasm to the nucleus. Co-localizes with ROCK2 in the nucleus.
Post-translational modifications:
Acetylated on Lys at up to 17 positions by intermolecular autocatalysis. Deacetylated in the transcriptional repression domain (CRD1) by SIRT1, preferentially at Lys-1020.
Citrullinated at Arg-2142 by PADI4, which impairs methylation by CARM1 and promotes interaction with NCOA2/GRIP1.
Methylated at Arg-580 and Arg-604 in the KIX domain by CARM1, which blocks association with CREB, inhibits CREB signaling and activates apoptotic response. Also methylated at Arg-2142 by CARM1, which impairs interaction with NCOA2/GRIP1.
Sumoylated; sumoylation in the transcriptional repression domain (CRD1) mediates transcriptional repression. Desumoylated by SENP3 through the removal of SUMO2 and SUMO3.
Probable target of ubiquitination by FBXO3, leading to rapid proteasome-dependent degradation.
Phosphorylated by HIPK2 in a RUNX1-dependent manner. This phosphorylation that activates EP300 happens when RUNX1 is associated with DNA and CBFB. Phosphorylated by ROCK2 and this enhances its activity. Phosphorylation at Ser-89 by AMPK reduces interaction with nuclear receptors, such as PPARG.
DISEASE:
Note=Defects in EP300 may play a role in epithelial cancer.
Note=Chromosomal aberrations involving EP300 may be a cause of acute myeloid leukemias. Translocation t(8;22)(p11;q13) with KAT6A.
Defects in EP300 are the cause of Rubinstein-Taybi syndrome type 2 (RSTS2) [MIM:613684]. A disorder characterized by craniofacial abnormalities, postnatal growth deficiency, broad thumbs, broad big toes, mental retardation and a propensity for development of malignancies. Some individuals with RSTS2 have less severe mental impairment, more severe microcephaly, and a greater degree of changes in facial bone structure than RSTS1 patients.
SWISS:
Q09472
Gene ID:
2033
Database links:Entrez Gene: 2033 Human
Entrez Gene: 328572 Mouse
Omim: 602700 Human
SwissProt: Q09472 Human
SwissProt: B2RWS6 Mouse
Unigene: 517517 Human
Unigene: 655211 Human
Unigene: 258397 Mouse
Unigene: 12447 Rat
Product Picture
Hela(Human) Cell Lysate at 30 ug
Primary: Anti- phospho-Ep300 (Ser1834) (SL5339R) at 1/1000 dilution
Secondary: IRDye800CW Goat Anti-Rabbit IgG at 1/20000 dilution
Predicted band size: 266 kD
Observed band size: 266 kD
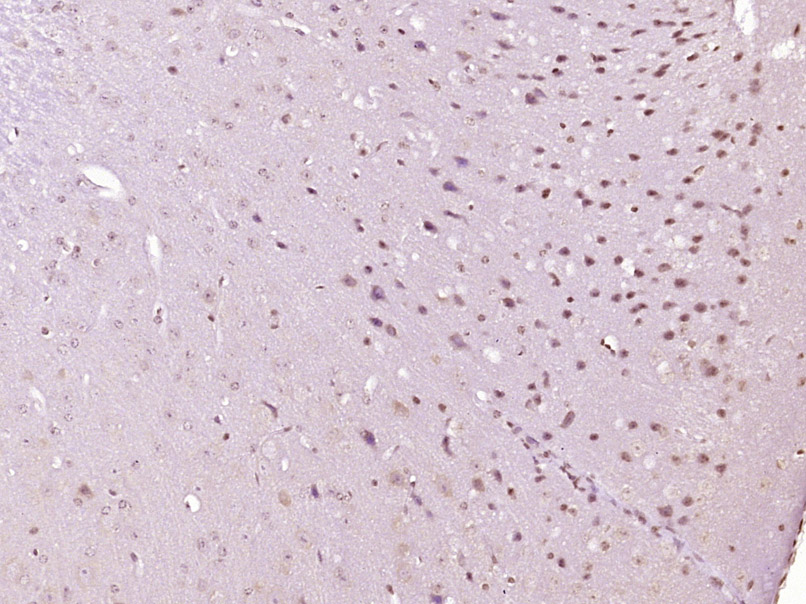
Paraformaldehyde-fixed, paraffin embedded (Mouse brain); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (phospho-Ep300 (Ser1834)) Polyclonal Antibody, Unconjugated (SL5339R) at 1:400 overnight at 4°C, followed by operating according to SP Kit(Rabbit) (sp-0023) instructionsand DAB staining.
Bought notes(bought amounts latest0)
No one bought this product
User Comment(Total0User Comment Num)
- No comment



 +86 571 56623320
+86 571 56623320
 +86 18668110335
+86 18668110335

