Rabbit Anti-EV71 polyprotein 3D antibody
3D polymerase; Human enterovirus 71 polyprotein 3D; EV71; Enterovirus 71; EV71 3D.
View History [Clear]
Details
Product Name EV71 polyprotein 3D Chinese Name 肠道病毒71型/手足口病病毒3D抗体 Alias 3D polymerase; Human enterovirus 71 polyprotein 3D; EV71; Enterovirus 71; EV71 3D. Research Area Cell biology immunology Bacteria and viruses Immunogen Species Rabbit Clonality Polyclonal Applications WB=1:500-2000 ELISA=1:5000-10000
not yet tested in other applications.
optimal dilutions/concentrations should be determined by the end user.Theoretical molecular weight 53/242kDa Form Liquid Concentration 1mg/ml immunogen KLH conjugated synthetic peptide derived from EV71 polyprotein 3D: 2101-2193/2193 Lsotype IgG Purification affinity purified by Protein A Buffer Solution 0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol. Storage Shipped at 4℃. Store at -20 °C for one year. Avoid repeated freeze/thaw cycles. Attention This product as supplied is intended for research use only, not for use in human, therapeutic or diagnostic applications. PubMed PubMed Product Detail Enteroviruses, such as enterovirus 71, are classified to be in the picornavirus family, pico [small] + RNA [ribonucleic acid] + virus. Picornaviruses are among the smallest and simplest ribonucleic acid containing viruses known (1). The RNA for many enteroviruses have now been cloned and complete genomic sequences have been obtained. The RNA from all sequenced enteroviruses are similar in length, about 7400 nucleotides, and have identical organization (1). The human alimentary tract is the predominant site of enterovirus replication and these viruses were first isolated from enteric specimens. These viruses are the cause of paralytic poliomyelitis, aseptic meningitis-encephalitis, myocarditis, pleurodynia, hand-foot-and-mouth disease, conjunctivitis, and numerous other syndromes associated with extra-intestinal target organs. There are 67 numbered types of enteroviruses in the enterovirus family (1): three polioviruses, twenty-three coxsackieviruses A, six coxsackieviruses B, thirty-one echoviruses, and four other enteroviruses.
Function:
Protein VP1: Forms, together with VP2 and VP3, an icosahedral capsid (pseudo T=3), 300 Angstroms in diameter, composed of 60 copies of each capsid protein and enclosing the viral positive strand RNA genome. Protein VP1 mainly forms the vertices of the capsid. VP1 interacts with host cell receptor to provide virion attachment to target cell. After binding to its receptor, the capsid undergoes conformational changes. VP1 N-terminus (that contains an amphipathic alpha-helix) is externalized, VP4 is released and together, they shape a virion-cell connecting channel and a pore in the host membrane through which RNase-protected transfer of the viral genome takes place. After genome has been released, the channel shrinks.
Protein VP2: Forms, together with VP1 and VP3, an icosahedral capsid (pseudo T=3), 300 Angstroms in diameter, composed of 60 copies of each capsid protein and
Protein VP3: Forms, together with VP1 and VP2, an icosahedral capsid (pseudo T=3), 300 Angstroms in diameter, composed of 60 copies of each capsid protein and enclosing the viral positive strand RNA genome.
Protein VP4: Lies on the inner surface of the capsid shell. After binding to the host receptor, the capsid undergoes conformational changes. VP4 is released, VP1 N-terminus is externalized, and together, they shape a virion-cell connecting channel and a pore in the host membrane through which RNase-protected transfer of the viral genome takes place. After genome has been released, the channel shrinks.
Protein VP0: Protein VP0: VP0 precursor is a component of immature procapsids, which gives rise to VP4 and VP2 afer maturation. Allows the capsid to remain inactive before the maturation step.
Protease 2A: cysteine protease that is responsible for the cleavage between the P1 and P2 regions. It cleaves the host translation initiation factor EIF4G1, in order to shut off the capped cellular mRNA transcription (By similarity).
Protein 2B: Affects membrane integrity and cause an increase in membrane permeability.
Protein 2C: Associates with and induces structural rearrangements of intracellular membranes. It displays RNA-binding, nucleotide binding and NTPase activities.
Protein 3A, via its hydrophobic domain, serves as membrane anchor. It also inhibits endoplasmic reticulum-to-Golgi transport.
Protease 3C: cysteine protease that generates mature viral proteins from the precursor polyprotein. In addition to its proteolytic activity, it binds to viral RNA, and thus influences viral genome replication. RNA and substrate bind cooperatively to the protease.
RNA-directed RNA polymerase 3D-POL replicates genomic and antigenomic RNA by recognizing replications specific signals.
Subunit:
Protein 2C N-terminus interacts with human RTN3. This interaction is important for viral replication.
Subcellular Location:
Protein VP2: Virion. Host cytoplasm (Potential).
Protein VP3: Virion. Host cytoplasm (Potential).
Protein VP1: Virion. Host cytoplasm (Potential).
Protein 2B: Host cytoplasmic vesicle membrane; Peripheral membrane protein; Cytoplasmic side (Potential). Note=Probably localizes to the surface of intracellular membrane vesicles that are induced after virus infection as the site for viral RNA replication. These vesicles are derived from the endoplasmic reticulum.
Protein 2C: Host cytoplasmic vesicle membrane; Peripheral membrane protein; Cytoplasmic side (Potential). Note=Probably localizes to the surface of intracellular membrane vesicles that are induced after virus infection as the site for viral RNA replication. These vesicles are derived from the endoplasmic reticulum.
Protein 3A: Host cytoplasmic vesicle membrane; Peripheral membrane protein; Cytoplasmic side (Potential). Note=Probably localizes to the surface of intracellular membrane vesicles that are induced after virus infection as the site for viral RNA replication. These vesicles are derived from the endoplasmic reticulum.
Protein 3B: Virion (Potential).
Protease 3C: Host cytoplasm (Potential).
RNA-directed RNA polymerase 3D-POL: Host cytoplasmic vesicle membrane; Peripheral membrane protein; Cytoplasmic side (Potential). Note=Probably localizes to the surface of intracellular membrane vesicles that are induced after virus infection as the site for viral RNA replication. These vesicles are derived from the endoplasmic reticulum.
Post-translational modifications:
Specific enzymatic cleavages in vivo by the viral proteases yield a variety of precursors and mature proteins. Polyprotein processing intermediates such as VP0 which is a VP4-VP2 precursor are produced. During virion maturation, non-infectious particles are rendered infectious following cleavage of VP0. This maturation cleavage is followed by a conformational change of the particle.
VPg is uridylylated by the polymerase and is covalently linked to the 5'-end of genomic RNA. This uridylylated form acts as a nucleotide-peptide primer for the polymerase.
Myristoylation of VP4 is required during RNA encapsidation and formation of the mature virus particle.
Similarity:
Belongs to the picornaviruses polyprotein family.
Contains 2 peptidase C3 domains.
Contains 1 RdRp catalytic domain.
Contains 1 SF3 helicase domain.
SWISS:
Q66478
Gene ID:
N/A
Database links:SwissProt: Q66478 HE71B
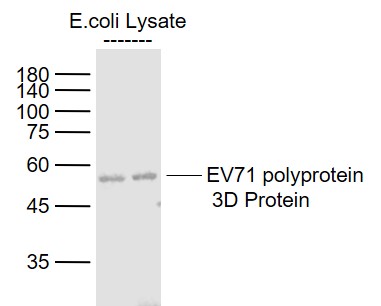
Product Picture
Partial purchase records(bought amounts latest0)
No one bought this product
User Comment(Total0User Comment Num)
- No comment


 +86 571 56623320
+86 571 56623320




