Rabbit Anti-MSH2 antibody
MSH-2; BAT26; DNA mismatch repair protein Msh2; FCC1; COCA1; HNPCC; LCFS2; HNPCC1; BAT26; COCA 1; COCA1; DNA mismatch repair protein Msh2; FCC 1; FCC1; hMSH2; HNPCC 1; HNPCC; HNPCC1; LCFS2; MSH 2; MSH2_HUMAN; MutS homolog 2; MutS homolog 2 colon cancer no
View History [Clear]
Details
Product Name MSH2 Chinese Name 错配修复蛋白2抗体 Alias MSH-2; BAT26; DNA mismatch repair protein Msh2; FCC1; COCA1; HNPCC; LCFS2; HNPCC1; BAT26; COCA 1; COCA1; DNA mismatch repair protein Msh2; FCC 1; FCC1; hMSH2; HNPCC 1; HNPCC; HNPCC1; LCFS2; MSH 2; MSH2_HUMAN; MutS homolog 2; MutS homolog 2 colon cancer nonpolyposis type 1; MutS protein homolog 2. literatures Research Area Tumour Cell biology Epigenetics Immunogen Species Rabbit Clonality Polyclonal React Species Human, Mouse, (predicted: Cow, ) Applications WB=1:500-2000 ELISA=1:5000-10000 IHC-F=1:100-500 IF=1:100-500 (Paraffin sections need antigen repair)
not yet tested in other applications.
optimal dilutions/concentrations should be determined by the end user.Theoretical molecular weight 105kDa Cellular localization The nucleus Form Liquid Concentration 1mg/ml immunogen KLH conjugated synthetic peptide derived from human MSH-2: 101-200/935 Lsotype IgG Purification affinity purified by Protein A Buffer Solution 0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol. Storage Shipped at 4℃. Store at -20 °C for one year. Avoid repeated freeze/thaw cycles. Attention This product as supplied is intended for research use only, not for use in human, therapeutic or diagnostic applications. PubMed PubMed Product Detail Component of the post-replicative DNA mismatch repair system (MMR). Forms two different heterodimers: MutS alpha (MSH2-MSH6 heterodimer) and MutS beta (MSH2-MSH3 heterodimer) which binds to DNA mismatches thereby initiating DNA repair. When bound, heterodimers bend the DNA helix and shields approximately 20 base pairs. MutS alpha recognizes single base mismatches and dinucleotide insertion-deletion loops (IDL) in the DNA. MutS beta recognizes larger insertion-deletion loops up to 13 nucleotides long. After mismatch binding, MutS alpha or beta forms a ternary complex with the MutL alpha heterodimer, which is thought to be responsible for directing the downstream MMR events, including strand discrimination, excision, and resynthesis. ATP binding and hydrolysis play a pivotal role in mismatch repair functions. The ATPase activity associated with MutS alpha regulates binding similar to a molecular switch: mismatched DNA provokes ADP-->ATP exchange, resulting in a discernible conformational transition that converts MutS alpha into a sliding clamp capable of hydrolysis-independent diffusion along the DNA backbone. This transition is crucial for mismatch repair. MutS alpha may also play a role in DNA homologous recombination repair. In melanocytes may modulate both UV-B-induced cell cycle regulation and apoptosis.
Function:
Component of the post-replicative DNA mismatch repair system (MMR). Forms two different heterodimers: MutS alpha (MSH2-MSH6 heterodimer) and MutS beta (MSH2-MSH3 heterodimer) which binds to DNA mismatches thereby initiating DNA repair. When bound, heterodimers bend the DNA helix and shields approximately 20 base pairs. MutS alpha recognizes single base mismatches and dinucleotide insertion-deletion loops (IDL) in the DNA. MutS beta recognizes larger insertion-deletion loops up to 13 nucleotides long. After mismatch binding, MutS alpha or beta forms a ternary complex with the MutL alpha heterodimer, which is thought to be responsible for directing the downstream MMR events, including strand discrimination, excision, and resynthesis. ATP binding and hydrolysis play a pivotal role in mismatch repair functions. The ATPase activity associated with MutS alpha regulates binding similar to a molecular switch: mismatched DNA provokes ADP-->ATP exchange, resulting in a discernible conformational transition that converts MutS alpha into a sliding clamp capable of hydrolysis-independent diffusion along the DNA backbone. This transition is crucial for mismatch repair. MutS alpha may also play a role in DNA homologous recombination repair. In melanocytes may modulate both UV-B-induced cell cycle regulation and apoptosis.
Subunit:
Heterodimer consisting of MSH2-MSH6 (MutS alpha) or MSH2-MSH3 (MutS beta). Both heterodimer form a ternary complex with MutL alpha (MLH1-PMS1). Interacts with EXO1. Part of the BRCA1-associated genome surveillance complex (BASC), which contains BRCA1, MSH2, MSH6, MLH1, ATM, BLM, PMS2 and the RAD50-MRE11-NBS1 protein complex. This association could be a dynamic process changing throughout the cell cycle and within subnuclear domains. Interacts with ATR. Interacts with SLX4/BTBD12; this interaction is direct and links MutS beta to SLX4, a subunit of different structure-specific endonucleases. Interacts with SMARCAD1.
Subcellular Location:
Nucleus.
Tissue Specificity:
Ubiquitously expressed.
Post-translational modifications:
Phosphorylated by PRKCZ, which may prevent MutS alpha degradation by the ubiquitin-proteasome pathway. Phosphorylated upon DNA damage, probably by ATM or ATR.
DISEASE:
Hereditary non-polyposis colorectal cancer 1 (HNPCC1) [MIM:120435]: An autosomal dominant disease associated with marked increase in cancer susceptibility. It is characterized by a familial predisposition to early-onset colorectal carcinoma (CRC) and extra-colonic tumors of the gastrointestinal, urological and female reproductive tracts. HNPCC is reported to be the most common form of inherited colorectal cancer in the Western world. Clinically, HNPCC is often divided into two subgroups. Type I is characterized by hereditary predisposition to colorectal cancer, a young age of onset, and carcinoma observed in the proximal colon. Type II is characterized by increased risk for cancers in certain tissues such as the uterus, ovary, breast, stomach, small intestine, skin, and larynx in addition to the colon. Diagnosis of classical HNPCC is based on the Amsterdam criteria: 3 or more relatives affected by colorectal cancer, one a first degree relative of the other two; 2 or more generation affected; 1 or more colorectal cancers presenting before 50 years of age; exclusion of hereditary polyposis syndromes. The term 'suspected HNPCC' or 'incomplete HNPCC' can be used to describe families who do not or only partially fulfill the Amsterdam criteria, but in whom a genetic basis for colon cancer is strongly suspected. Note=The disease is caused by mutations affecting the gene represented in this entry.
Muir-Torre syndrome (MRTES) [MIM:158320]: Rare autosomal dominant disorder characterized by sebaceous neoplasms and visceral malignancy. Note=The disease is caused by mutations affecting the gene represented in this entry.
Endometrial cancer (ENDMC) [MIM:608089]: A malignancy of endometrium, the mucous lining of the uterus. Most endometrial cancers are adenocarcinomas, cancers that begin in cells that make and release mucus and other fluids. Note=Disease susceptibility is associated with variations affecting the gene represented in this entry.
Similarity:
Belongs to the DNA mismatch repair MutS family.
SWISS:
P43246
Gene ID:
4436
Database links:Entrez Gene: 4436 Human
Entrez Gene: 17685 Mouse
Omim: 120435 Human
SwissProt: P43246 Human
SwissProt: P43247 Mouse
Unigene: 597656 Human
Unigene: 4619 Mouse
Unigene: 471165 Mouse
Unigene: 3174 Rat
MSH2是核苷酸错配修复酶之一,其突变和多种Tumour的发生有关,主要用于:先天性结肠癌、散发性结肠癌及一些消化系统Tumour方面的研究。DNA错配修复系统是指由特异修复DNA碱基错配的酶组成,保证DNA遗传物质的高保真性。它由MLH1、MSH2、PMS1、PMS2、MSH6和MSH3基因组成。如果上述基因发生突变或失活,会使细胞错配修复功能缺陷,产生遗传不稳定性,导致Tumour易感。MSH2基因突变发生在散发性结肠癌中。
hMSH-2和hMLH1都与Tumour的发生有关。Product Picture
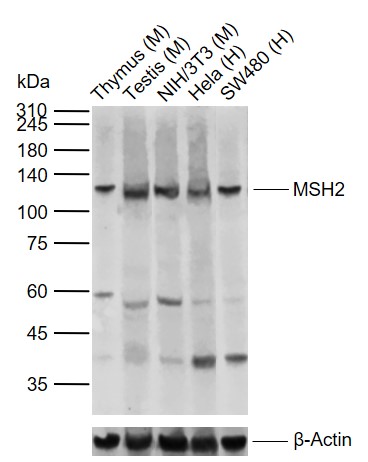
Lane 1: Mouse Thymus tissue lysates
Lane 2: Mouse Testis tissue lysates
Lane 3: Mouse NIH/3T3 cell lysates
Lane 4: Human Hela cell lysates
Lane 5: Human SW480 cell lysates
Primary: Anti-MSH2 (SL0758R) at 1/1000 dilution
Secondary: IRDye800CW Goat Anti-Rabbit IgG at 1/20000 dilution
Predicted band size: 105 kDa
Observed band size: 110 kDa
Bought notes(bought amounts latest0)
No one bought this product
User Comment(Total0User Comment Num)
- No comment


 +86 571 56623320
+86 571 56623320
 +86 18668110335
+86 18668110335

