Mouse Anti-CTCF antibody
11 zinc finger protein; 11 zinc finger transcriptional repressor; CCCTC binding factor (zinc finger protein); CCCTC binding factor; CTCFL paralog; Transcriptional repressor CTCF; CTCF_HUMAN.
View History [Clear]
Details
Product Name CTCF Chinese Name 转录阻抑蛋白CTCF单克隆抗体 Alias 11 zinc finger protein; 11 zinc finger transcriptional repressor; CCCTC binding factor (zinc finger protein); CCCTC binding factor; CTCFL paralog; Transcriptional repressor CTCF; CTCF_HUMAN. Research Area Tumour Cell biology Developmental biology Chromatin and nuclear signals transcriptional regulatory factor Immunogen Species Mouse Clonality Monoclonal Clone NO. G3F32 React Species Human, Rat, Applications WB=1:500-2000
not yet tested in other applications.
optimal dilutions/concentrations should be determined by the end user.Theoretical molecular weight 83kDa Cellular localization The nucleus Form Liquid Concentration 1mg/ml immunogen Recombinant human CTCF between 445-727 amino acids. Lsotype IgG1,k Purification affinity purified by Protein G Buffer Solution 0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol. Storage Shipped at 4℃. Store at -20 °C for one year. Avoid repeated freeze/thaw cycles. Attention This product as supplied is intended for research use only, not for use in human, therapeutic or diagnostic applications. PubMed PubMed Product Detail CTCF (CCCTC-binding factor) is a highly conserved zinc finger protein that has been implicated in playing a role in a wide range of regulatory functions, including transcriptional activation/repression, insulation, imprinting, and X chromosome inactivation. A general role for CTCF in the global organisation of chromatin architecture has been proposed. It has been suggested that CTCF is involved, in a heritable manner, in the interplay between DNA methylation, higher-order chromatin structure, and lineage-specific gene expression.
Function:
Chromatin binding factor that binds to DNA sequence specific sites. Involved in transcriptional regulation by binding to chromatin insulators and preventing interaction between promoter and nearby enhancers and silencers. Acts as transcriptional repressor binding to promoters of vertebrate MYC gene and BAG1 gene. Also binds to the PLK and PIM1 promoters. Acts as a transcriptional activator of APP. Regulates APOA1/C3/A4/A5 gene cluster and controls MHC class II gene expression. Plays an essential role in oocyte and preimplantation embryo development by activating or repressing transcription. Seems to act as tumor suppressor. Plays a critical role in the epigenetic regulation. Participates in the allele-specific gene expression at the imprinted IGF2/H19 gene locus. On the maternal allele, binding within the H19 imprinting control region (ICR) mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to IGF2. Plays a critical role in gene silencing over considerable distances in the genome. Preferentially interacts with unmethylated DNA, preventing spreading of CpG methylation and maintaining methylation-free zones. Inversely, binding to target sites is prevented by CpG methylation. Plays a important role in chromatin remodeling. Can dimerize when it is bound to different DNA sequences, mediating long-range chromatin looping. Mediates interchromosomal association between IGF2/H19 and WSB1/NF1 and may direct distant DNA segments to a common transcription factory. Causes local loss of histone acetylation and gain of histone methylation in the beta-globin locus, without affecting transcription. When bound to chromatin, it provides an anchor point for nucleosomes positioning. Seems to be essential for homologous X-chromosome pairing. May participate with Tsix in establishing a regulatable epigenetic switch for X chromosome inactivation. May play a role in preventing the propagation of stable methylation at the escape genes from X- inactivation. Involved in sister chromatid cohesion. Associates with both centromeres and chromosomal arms during metaphase and required for cohesin localization to CTCF sites. Regulates asynchronous replication of IGF2/H19.
Subunit:
Interacts with CHD8.
Subcellular Location:
Nucleus, nucleoplasm. Chromosome. Chromosome, centromere. Note=May translocate to the nucleolus upon cell differentiation. Associates with both centromeres and chromosomal arms during metaphase. Associates with the H19 ICR in mitotic chromosomes. May be preferentially excluded from heterochromatin during interphase.
Tissue Specificity:
Ubiquitous. Absent in primary spermatocytes.
Post-translational modifications:
Sumoylated on Lys-74 and Lys-689; sumoylation of CTCF contributes to the repressive function of CTCF on the MYC P2 promoter.
Similarity:
Belongs to the CTCF zinc-finger protein family. Contains 11 C2H2-type zinc fingers.
SWISS:
P49711
Gene ID:
10664
Database links:Entrez Gene: 10664 Human
SwissProt: P49711 Human
Product Picture
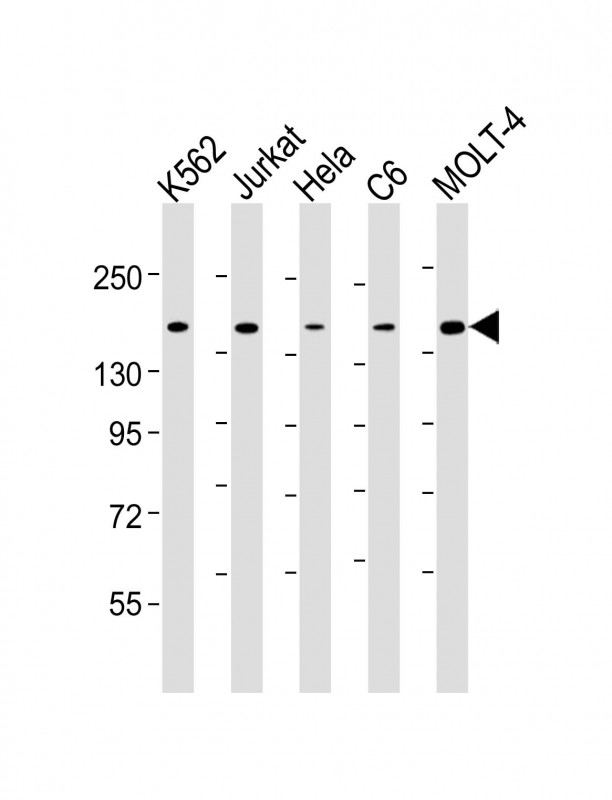
Lane 1: K562 cell lysates
Lane 2: Jurkat cell lysates
Lane 3: Hela cell lysates
Lane 4: C6 cell lysates
Lane 5: MOLT-4 cell lysates
Primary: Anti-CTCF (SLM-51632M) at 1/4000 dilution
Secondary: IRDye800CW Goat Anti-Mouse IgG at 1/20000 dilution
Predicted band size: 83 kD
Observed band size: 180 kD
References (0)
No References
Bought notes(bought amounts latest0)
No one bought this product
User Comment(Total0User Comment Num)
- No comment


 +86 571 56623320
+86 571 56623320
 +86 18668110335
+86 18668110335

